Automating Computational Reproducibility in R using renv, Docker, and GitHub Actions
The Problem
Have you ever wanted to run someone else’s R code (or your own—from a distant past), but upon downloading it, you realized that it relied on versions of either R or R libraries that are dramatically different from those on your own computer? This situation can easily spiral into dependency hell, a place that every researcher or data scientist will (always unintentionally) find themselves when trying to get random code from bygone days to run locally. Oftentimes, it may seem impossible to resolve The Problem, and giving up may seem like the only logical thing to do…

Caption: Occasionally, dependency hell peak’s its head into the real world too
The Naive Attempt to Resolve The Problem
If you are like me, the first thing you might do when finding yourself in dependency hell is to try installing different versions of the R libraries that R is complaining about. If that doesn’t work, you may even get desperate enough to try installing a different version of R in hopes that The Problem will magically disappear.
In some cases, the naive attempt can succeed. But alas, it will often fail, and even when it does succeed—that different version of R (or R libraries) you installed could lead to problems in other ongoing projects. You know, the projects that you briefly forgot about while trying to resolve The Problem for the current project on the to-do list. In this case, dependency hell has evolved into computational whac-a-mole—a rage-inducing game that your therapist told you to stop playing many sessions ago.

Caption: You, disregarding your therapist
A Slightly Better Attempt, But It Requires Foresight
If you truly want to resolve dependency issues, you will need to think ahead and build some form of dependency management into your workflow (I know, very unfortunate…). That said, it is getting easier with each passing year to automate a good deal of it. In the remainder of this post, we will walk through how to create a GitHub repository that takes advantage of renv, Docker, and GitHub Actions to make your R workflow reproducible into the future.
(NOTE: Throughout this post, you can refer to my accompanying GitHub repo to see all the code in one place. I would recommend forking it, cloning to a local directory, and testing things out that way. Then, you can try out some of your own R scripts to see how things work!)
An Overview of renv
renv is an R package that allows for us to install project-specific packages as opposed to relying on the same set of packages (and their associated versions) across all R projects. Further, we can take a snapshot of the packages we are using in a given project, save the snapshot for later, and then restore (i.e. reinstall) the packages later—even on different computers.
Starting a New Project with renv
Using renv is very easy. First, while in R, set your working directory to the folder that we want to create our project in:
# This is the GitHub repo associated with this project
setwd("~/computational-reproducibility-in-r")Next, install renv:
install.packages("renv")Now, we simply load the package and then initialize the project:
renv::init()
# * Initializing project ...
# * Discovering package dependencies ... Done!
# * Copying packages into the cache ... Done!
# * Lockfile written to # '~/computational-reproducibility-in-r/renv.lock'.
# Restarting R session...Installing Packages with renv
With the project initialized, we essentially proceed as normal, except that we will install all the packages we need along the way. For example, if our project uses dplyr and ggplot2, we install them as follows:
install.packages(c("dplyr", "ggplot2"))
# Installing dplyr [1.0.7] ...
# OK [linked cache]
# Installing ggplot2 [3.3.5] ...
# OK [linked cache]Now, both packages are available to use in our project. Let’s make a plot for proof!
library(dplyr)
library(ggplot2)
set.seed(43210)
# Yes, your love grows exponentially
love_for_renv <- rnorm(100, exp(.022*(1:100)), .8)
data.frame(Time = 1:100,
love_for_renv = love_for_renv) %>%
ggplot(aes(x = Time, y = love_for_renv)) +
geom_point() +
geom_line() +
ylab("Love for renv") +
scale_y_continuous(breaks = 1:10, limits = c(0,11)) +
theme_minimal(base_size = 15) +
theme(panel.grid = element_blank())Now, let’s say that we are wrapping up for the day and would like to push our changes to GitHub. After saving our R file that produces the beautiful graph above, we need to take a snapshot so that renv remembers which libraries we have installed for the current project:
renv::snapshot()
# The following package(s) will be updated in the lockfile:
#
# # CRAN ===============================
# - MASS [* -> 7.3-54]
# - Matrix [* -> 1.3-3]
# - R6 [* -> 2.5.1]
# - RColorBrewer [* -> 1.1-2]
# - cli [* -> 3.0.1]
# - colorspace [* -> 2.0-2]
# - crayon [* -> 1.4.1]
# - digest [* -> 0.6.29]
# - dplyr [* -> 1.0.7]
# - ellipsis [* -> 0.3.2]
# - fansi [* -> 0.5.0]
# - farver [* -> 2.1.0]
# - generics [* -> 0.1.0]
# - ggplot2 [* -> 3.3.5]
# - glue [* -> 1.6.0]
# - gtable [* -> 0.3.0]
# - isoband [* -> 0.2.5]
# - labeling [* -> 0.4.2]
# - lattice [* -> 0.20-44]
# - lifecycle [* -> 1.0.0]
# - magrittr [* -> 2.0.1]
# - mgcv [* -> 1.8-35]
# - munsell [* -> 0.5.0]
# - nlme [* -> 3.1-152]
# - pillar [* -> 1.6.2]
# - pkgconfig [* -> 2.0.3]
# - purrr [* -> 0.3.4]
# - rlang [* -> 0.4.12]
# - scales [* -> 1.1.1]
# - tibble [* -> 3.1.4]
# - tidyselect [* -> 1.1.1]
# - utf8 [* -> 1.2.2]
# - vctrs [* -> 0.3.8]
# - viridisLite [* -> 0.4.0]
# - withr [* -> 2.4.2]
#
# Do you want to proceed? [y/N]: y
# * Lockfile written to
# '~/computational-reproducibility-in-r/renv.lock'.That’s a lot of dependencies for just two R packages… which is exactly why we want to use something like renv to automate version tracking for us! Note that renv searches through the files in our project directory to determine which packages we are using, so make sure that you have a file with your R code saved somewhere in your project directory before taking the snapshot.
If you look through your project directory, you will notice that renv has created a few different files. Of these, the renv.lock file is what tracks the packages and their associated versions that your project depends on. Also, you will see a line in your .Rprofile file (which may be hidden, in which case you will need to show the hidden files to see it; e.g., in a terminal, run ls -la) that looks like: source("renv/activate.R"). If you did not know, this file is run each time R is started from your project directory. In this case, it activates renv automatically for you, making all the packages available for the current project. This way, if you were to open your project on another computer, you would have all the correct packages/versions ready to go. Simply running renv::restore() will re-install all of them, after which you can proceed as normal. Pretty cool, right?
And that is pretty much it for renv! However, you may have recognized a problem—even if we have the right package versions, what if we want to open our project on a computer with a different version of R? Or, what if we share our project with someone on a different operating system? In these cases, just having the correct versions for our packages is not enough—we need the correct R version too. This is where Docker comes in…
Packaging up your R Project with Docker
Docker is important because it allows us to package up not only our packages, but also our R version and even our operating system! In this way, creating a Docker image will ensure that (almost) any computer with Docker installed will be able to run our code. This is particularly useful when we want to run computationally intensive code on cloud services such as AWS, GCP, or others.
The Basic Steps in Building a Docker Image
After installing Docker (see instructions here), building an image is rather straightforward—at least, it is with a bit of practice! There are basically three key steps involved in building a Docker image:
- Specifying the starting image to build off of,
- Installing the necessary dependencies/packages that you want to use, and
- Telling the image how to start when you run it
For us, we want our image to start using some basic version of R that matches the version we have on our computer when developing a project. Then, we want to install all the R packages that are specified in the renv.lock file described earlier. Finally, we have a choice to either make the image start up R right away, or we can alternatively start from a terminal. I like to start from the terminal, personally, but this is just preference.
To follow the above steps, we can first determine our R version by running R --version in the terminal. In my case, I currently have version 4.1.0 installed:
# R version 4.1.0 (2021-05-18) -- "Camp Pontanezen"
# Copyright (C) 2021 The R Foundation for Statistical Computing
# Platform: aarch64-apple-darwin20 (64-bit)
#
# R is free software and comes with ABSOLUTELY NO WARRANTY.
# You are welcome to redistribute it under the terms of the
# GNU General Public License versions 2 or 3.
# For more information about these matters see
# https://www.gnu.org/licenses/.Next, we need to know our version of renv too. For this, you can just look at the renv.lock file and search for renv. With these two pieces of information, we can create a Dockerfile (a raw text file named “Dockerfile” with no file extensions, located in your project root directory), which is what is used to build a Docker image. Here is the Dockerfile that I use for the current example project:
# Start with R version 4.1.0
FROM rocker/r-ver:4.1.0
# Install some linux libraries that R packages need
RUN apt-get update && apt-get install -y libcurl4-openssl-dev libssl-dev libxml2-dev
# Use renv version 0.15.2
ENV RENV_VERSION 0.15.2
# Install renv
RUN Rscript -e "install.packages('remotes', repos = c(CRAN = 'https://cloud.r-project.org'))"
RUN Rscript -e "remotes::install_github('rstudio/renv@${RENV_VERSION}')"
# Create a directory named after our project directory
WORKDIR /computational-reproducibility-in-r
# Copy the lockfile over to the Docker image
COPY renv.lock renv.lock
# Install all R packages specified in renv.lock
RUN Rscript -e 'renv::restore()'
# Default to bash terminal when running docker image
CMD ["bash"]The comments above detail what is going on—for you, you can make this work for your project by simply changing the R and renv version numbers to the values for your own project.
To actually build the image so that we can run it, we can do so in the terminal using the following code:
docker build -t my-docker-image .
# [+] Building 150.2s (11/11) FINISHED
# => [internal] load build definition from Dockerfile 0.0s
# => => transferring dockerfile: 694B 0.0s
# => [internal] load .dockerignore 0.0s
# => => transferring context: 2B 0.0s
# => [internal] load metadata for docker.io/rocker/r-ver:4.1.0 0.0s
# => [internal] load build context 0.0s
# => => transferring context: 9.10kB 0.0s
# => CACHED [1/6] FROM docker.io/rocker/r-ver:4.1.0 0.0s
# => [2/6] RUN Rscript -e "install.packages('remotes', repos = c(CRAN = 'h 5.9s
# => [3/6] RUN Rscript -e "remotes::install_github('rstudio/renv@0.15.2') 12.8s
# => [4/6] WORKDIR /computational-reproducibility-in-r 0.0s
# => [5/6] COPY renv.lock renv.lock 0.0s
# => [6/6] RUN Rscript -e 'renv::restore()' 130.7s
# => exporting to image 0.7s
# => => exporting layers 0.7s
# => => writing image sha256:348a67e88477b16e01bd38a34e73c1c72c2f12e9ec927 0.0s
# => => naming to docker.io/library/my-docker-image 0.0s The code above tells Docker to build the image and tag it with the name my-docker-image. Looks like it worked!
Note: You may be wondering—where are those random linux libraries in the Dockerfile coming from? How did you know you needed them? Well, I didn’t actually know when I started! When I tried building the image without them, I would get an error message that pointed toward a library that was needed. For example, here is one of the error messages:
#9 628.1 Installing xml2 [1.3.2] ...
#9 628.3 FAILED
#9 628.3 Error installing package 'xml2':
#9 628.3 ================================
#9 628.3
#9 628.3 * installing to library ‘/usr/local/lib/R/site-library/.renv/1’
#9 628.3 * installing *source* package ‘xml2’ ...
#9 628.3 ** package ‘xml2’ successfully unpacked and MD5 sums checked
#9 628.3 ** using staged installation
#9 628.3 Using PKG_CFLAGS=
#9 628.3 Using PKG_LIBS=-lxml2
#9 628.3 ------------------------- ANTICONF ERROR ---------------------------
#9 628.3 Configuration failed because libxml-2.0 was not found. Try installing:
#9 628.3 * deb: libxml2-dev (Debian, Ubuntu, etc)
#9 628.3 * rpm: libxml2-devel (Fedora, CentOS, RHEL)
#9 628.3 * csw: libxml2_dev (Solaris)
#9 628.3 If libxml-2.0 is already installed, check that 'pkg-config' is in your
#9 628.3 PATH and PKG_CONFIG_PATH contains a libxml-2.0.pc file. If pkg-config
#9 628.3 is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
#9 628.3 R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...'
#9 628.3 --------------------------------------------------------------------
#9 628.3 ERROR: configuration failed for package ‘xml2’
#9 628.3 * removing ‘/usr/local/lib/R/site-library/.renv/1/xml2’
#9 628.3 Error: install of package 'xml2' failed [error code 1]This is a really helpful message—it says that libxml2-dev is necessary for installing the R package xml2, and because our image did not have libxml2-dev, the build failed. So, by adding libxml2-dev in the Dockerfile, the problem was resolved. The same thing happened for the other libraries in the Dockerfile. So, if you run into this issue, just check out the error message to find what you need, and add it to the line in the Dockerfile.
Running a Docker Image
To actually be of any use, we need to be able to run code that is in our project using our Docker image. To do so, it is important to note that when you run a Docker image, it is completely self-contained (which is why it is called a Docker container when you are running it). This means that you will not have access to any of your local files when you are running your container unless you connect the container to your local directory.
To do this, we can use the -v flag when running Docker:
docker run -it -v ~/computational-reproducibility-in-r:/computational-reproducibility-in-r my-docker-imageAbove, docker run is starting the container, -it is making it interactive mode (so that we can type out commands ourselves), and the <path-to-my-local-project-directory>:<docker-directory-to-mount-to> part tells Docker to mount our local project directory to the computational-reproducibility-in-r directory that we created in the Dockerfile above (see the line that reads WORKDIR /computational-reproducibility-in-r). When mounted, anything we save to the mounted directory will be saved to our local directory too! So, if we create an R script called make_beautiful_plot.R that contains the same plotting code from above, but it saves the image to our local directory instead of just showing it:
library(dplyr)
library(ggplot2)
set.seed(43210)
love_for_renv <- rnorm(100, exp(.022*(1:100)), .8)
my_plot <-
data.frame(Time = 1:100,
love_for_renv = love_for_renv) %>%
ggplot(aes(x = Time, y = love_for_renv)) +
geom_point() +
geom_line() +
ylab("Love for renv") +
scale_y_continuous(breaks = 1:10, limits = c(0,11)) +
theme_minimal(base_size = 15) +
theme(panel.grid = element_blank())
ggsave(my_plot, file = "saved_plot.pdf", unit = "in",
height = 4, width = 5)we can run this script within the Docker container by running:
Rscript make_beautiful_plot.Rand we will see saved_plot.pdf appear in our local project directory!
Although we just made a basic plot, what we have achieved is rather cool—theoretically, anyone could use the image that we created to run our code, making our analyses computationally reproducible across time and across different computers.
But… there is one issue… how do we share the image? Also, building the image manually and then sending it somewhere seems like a big pain if I am working on a project over the course of months or longer. Well, this is where GitHub and GitHub Actions come to play!
Using GitHub to Build and Store Docker Images
GitHub is generally an amazing platform for version control when it comes to code, but it can also play the same role for our Docker images. Further, we can use GitHub Actions to automate a good deal of our workflow—in our case, to build and push our Docker images to a repository that allows for anyone to access them.
To start, once you have your local project configured to be linked with a GitHub repo, as long as you have a Dockerfile in your project directory, you can actually use the template provided by GitHub to create a workflow for building and pushing images to your own GitHub account:
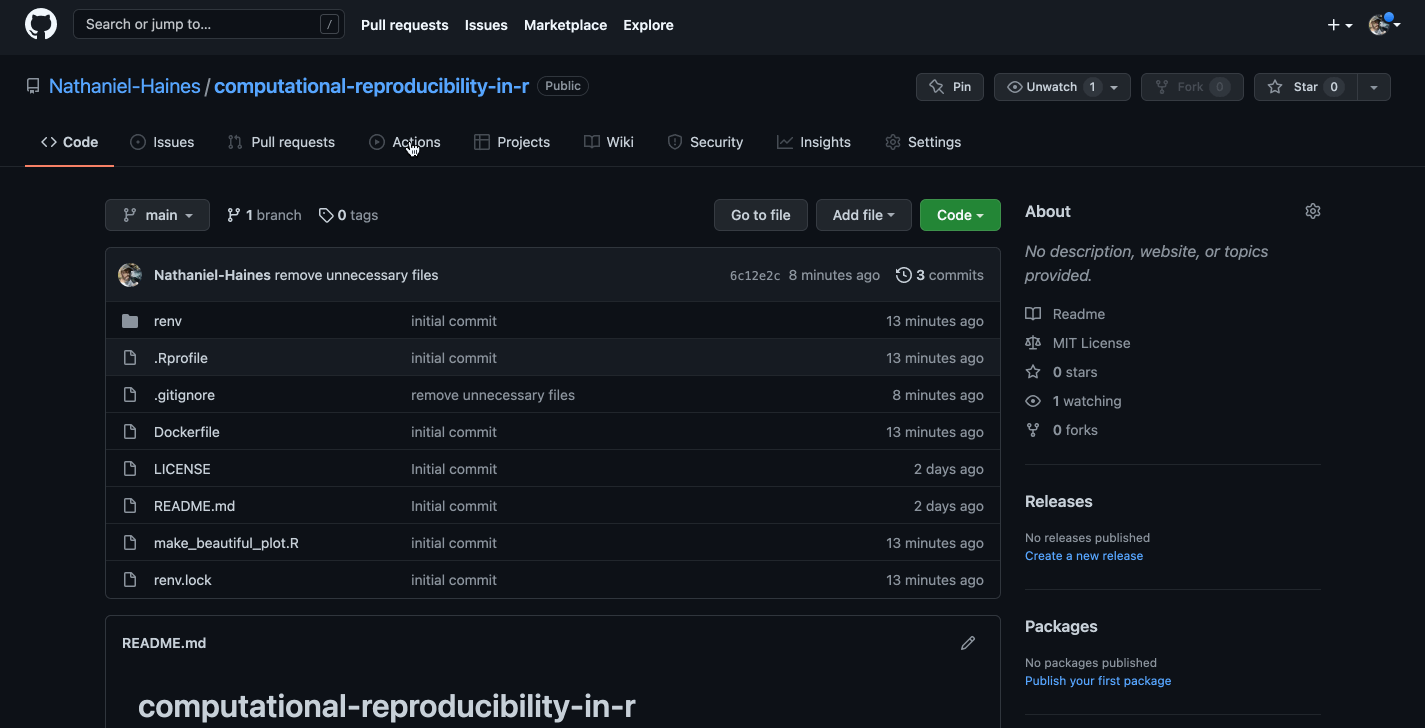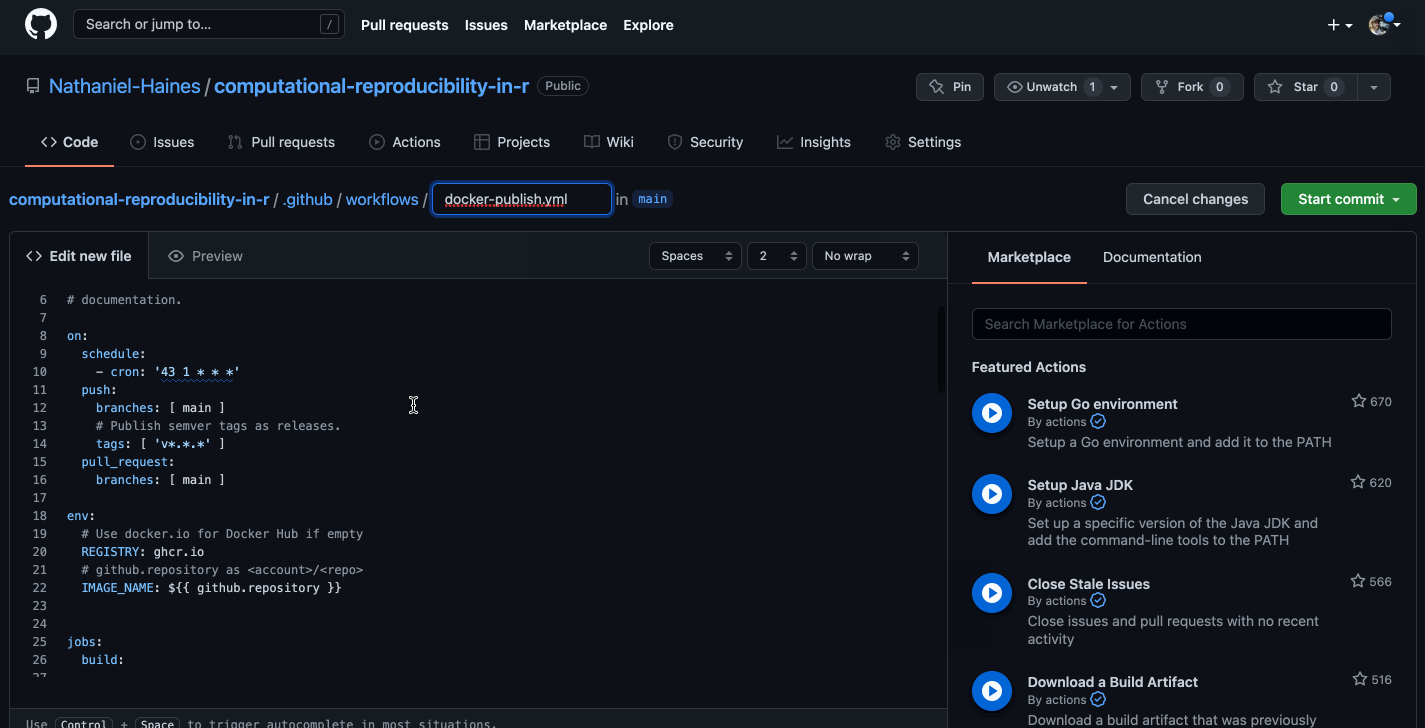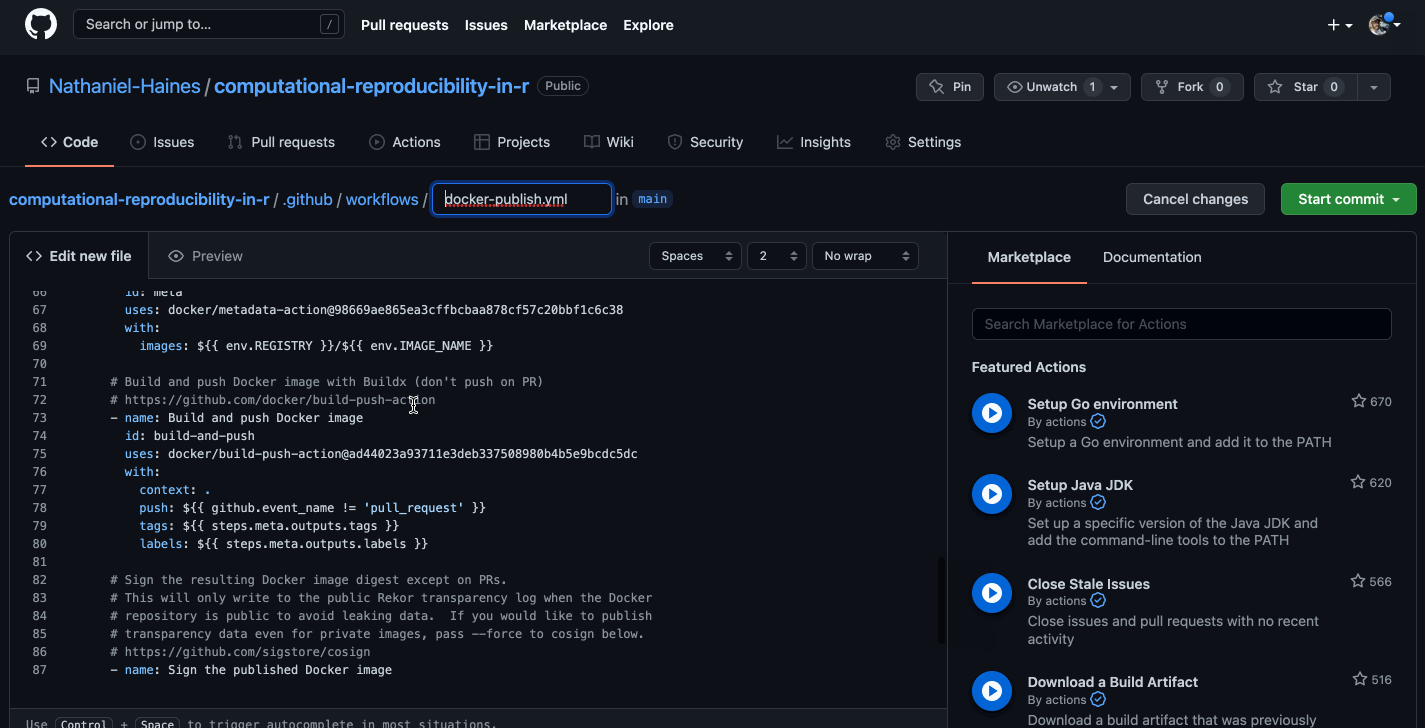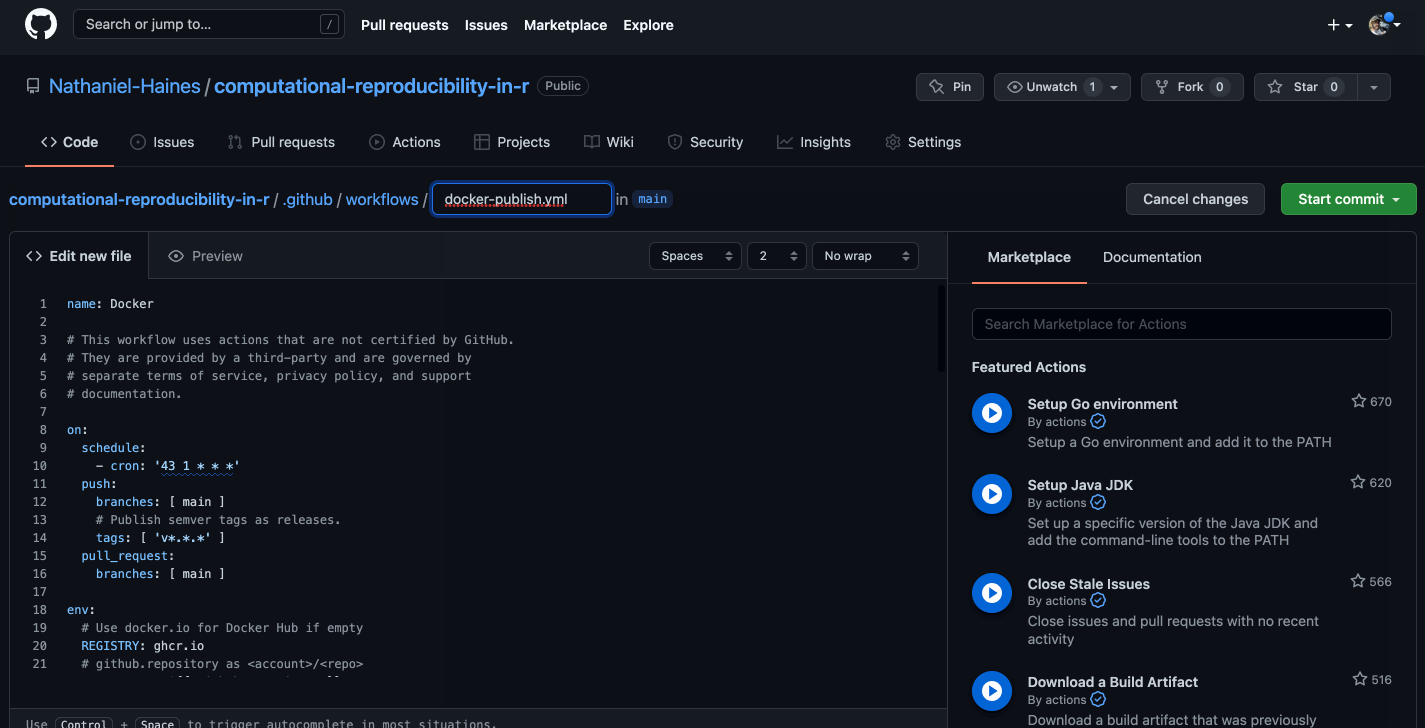
Caption: Well, that looks a bit confusing…
That said, the template didn’t make a whole lot of sense to me at first, so I opted to follow the documentation here in hopes that I would better understand the final product.
The basic idea is that we want to build a new image using the current dependencies in our project each time that we push changes to GitHub. I made some changes the the linked code above that make this happen. The resulting code is here (I would recommend checking out the link for more details on how this works):
name: Docker
# This workflow uses actions that are not certified by GitHub.
# They are provided by a third-party and are governed by
# separate terms of service, privacy policy, and support
# documentation.
on:
push:
branches: [ main ]
# Publish semver tags as releases.
tags: [ 'v*.*.*' ]
env:
# Use docker.io for Docker Hub if empty
REGISTRY: ghcr.io
# github.repository as <account>/<repo>
IMAGE_NAME: ${{ github.repository }}
jobs:
# Push image to GitHub Packages.
# See also https://docs.docker.com/docker-hub/builds/
push:
runs-on: ubuntu-latest
permissions:
packages: write
contents: read
steps:
- uses: actions/checkout@v2
with:
fetch-depth: 0 # OR "2" -> To retrieve the preceding commit.
- name: Run file diff checking image
uses: tj-actions/changed-files@v14.1
- name: Build image
if: contains(steps.changed-files.outputs.modified_files, 'renv.lock')
run: |
IMAGE_NAME=$(echo $IMAGE_NAME | tr '[A-Z]' '[a-z]')
docker build . --file Dockerfile --tag $IMAGE_NAME --label "runnumber=${GITHUB_RUN_ID}"
- name: Log in to registry
# This is where you will update the PAT to GITHUB_TOKEN
run: echo "${{ secrets.GITHUB_TOKEN }}" | docker login ghcr.io -u ${{ github.actor }} --password-stdin
- name: Push image
if: contains(steps.changed-files.outputs.modified_files, 'renv.lock')
run: |
IMAGE_ID=ghcr.io/$IMAGE_NAME
# Change all uppercase to lowercase
IMAGE_ID=$(echo $IMAGE_ID | tr '[A-Z]' '[a-z]')
IMAGE_NAME=$(echo $IMAGE_NAME | tr '[A-Z]' '[a-z]')
# Strip git ref prefix from version
VERSION=$(echo "${{ github.ref }}" | sed -e 's,.*/\(.*\),\1,')
# Strip "v" prefix from tag name
[[ "${{ github.ref }}" == "refs/tags/"* ]] && VERSION=$(echo $VERSION | sed -e 's/^v//')
# Use Docker `latest` tag convention
[ "$VERSION" == "main" ] && VERSION=latest
echo IMAGE_ID=$IMAGE_ID
echo VERSION=$VERSION
docker tag $IMAGE_NAME $IMAGE_ID:$VERSION
docker push $IMAGE_ID:$VERSIONNote that, as in the template shown in the .gif above, the code chunk above is saved in computational-reproducibility-in-r/.github/workflows/docker-publish.yml. Once this file is in your GitHub repository, all we need to do is make changes to our code and push them to GitHub, and a new Docker image will be built and pushed to your GitHub Packages registry (which we will walk through later) automatically! Let’s test it out.
Testing Out Our Complete Workflow
To test things out, let’s install a new R package and then push our updated code to GitHub. This will trigger the GitHub Action from above, which will then build and push an image to our GitHub Packages registry. First, let’s install the tidyverse. After opening an R terminal in our project directory:
install.packages("tidyverse")
# Retrieving 'https://cran.rstudio.com/bin/macosx/big-sur-arm64/contrib/4.1/dtplyr_1.2.1.tgz' ...
# OK [downloaded 321 Kb in 0.3 secs]
# Retrieving 'https://cran.rstudio.com/bin/macosx/big-sur-arm64/contrib/4.1/rvest_1.0.2.tgz' ...
# OK [downloaded 195 Kb in 0.3 secs]
# Installing dtplyr [1.2.1] ...
# OK [installed binary]
# Moving dtplyr [1.2.1] into the cache ...
# OK [moved to cache in 0.43 milliseconds]
# Installing forcats [0.5.1] ...
# OK [linked cache]
# Installing googledrive [2.0.0] ...
# OK [linked cache]
# Installing rematch [1.0.1] ...
# OK [linked cache]
# Installing cellranger [1.1.0] ...
# OK [linked cache]
# Installing ids [1.0.1] ...
# OK [linked cache]
# Installing googlesheets4 [1.0.0] ...
# OK [linked cache]
# Installing haven [2.4.3] ...
# OK [linked cache]
# Installing modelr [0.1.8] ...
# OK [linked cache]
# Installing readxl [1.3.1] ...
# OK [linked cache]
# Installing reprex [2.0.1] ...
# OK [linked cache]
# Installing selectr [0.4-2] ...
# OK [linked cache]
# Installing rvest [1.0.2] ...
# OK [installed binary]
# Moving rvest [1.0.2] into the cache ...
# OK [moved to cache in 0.47 milliseconds]
# Installing tidyverse [1.3.1] ...
# OK [linked cache]You will see renv do its magic, as per usual. Then, we need to modify our code to include the tidyverse so that renv can detect it when taking a new snapshot. I will just add a new file called new_dependency.R that only runs library(tidyverse) as an example. After creating this file, we go back to an R terminal and run:
renv::snapshot()
# The following package(s) will be updated in the lockfile:
#
# # CRAN ===============================
# - DBI [* -> 1.1.1]
# - Rcpp [* -> 1.0.8]
# - askpass [* -> 1.1]
# - assertthat [* -> 0.2.1]
# - backports [* -> 1.2.1]
# - base64enc [* -> 0.1-3]
# - bit [* -> 4.0.4]
# - bit64 [* -> 4.0.5]
# - blob [* -> 1.2.2]
# - broom [* -> 0.7.11]
# - callr [* -> 3.7.0]
# - cellranger [* -> 1.1.0]
# - clipr [* -> 0.7.1]
# - cpp11 [* -> 0.4.1]
# - curl [* -> 4.3.2]
# - data.table [* -> 1.14.0]
# - dbplyr [* -> 2.1.1]
# - dtplyr [* -> 1.2.1]
# - evaluate [* -> 0.14]
# - fastmap [* -> 1.1.0]
# - forcats [* -> 0.5.1]
# - fs [* -> 1.5.0]
# - gargle [* -> 1.2.0]
# - googledrive [* -> 2.0.0]
# - googlesheets4 [* -> 1.0.0]
# - haven [* -> 2.4.3]
# - highr [* -> 0.9]
# - hms [* -> 1.1.0]
# - htmltools [* -> 0.5.2]
# - httr [* -> 1.4.2]
# - ids [* -> 1.0.1]
# - jquerylib [* -> 0.1.4]
# - jsonlite [* -> 1.7.3]
# - knitr [* -> 1.37]
# - lubridate [* -> 1.8.0]
# - mime [* -> 0.12]
# - modelr [* -> 0.1.8]
# - openssl [* -> 1.4.5]
# - prettyunits [* -> 1.1.1]
# - processx [* -> 3.5.2]
# - progress [* -> 1.2.2]
# - ps [* -> 1.6.0]
# - rappdirs [* -> 0.3.3]
# - readr [* -> 2.0.2]
# - readxl [* -> 1.3.1]
# - rematch [* -> 1.0.1]
# - rematch2 [* -> 2.1.2]
# - reprex [* -> 2.0.1]
# - rmarkdown [* -> 2.11]
# - rstudioapi [* -> 0.13]
# - rvest [* -> 1.0.2]
# - selectr [* -> 0.4-2]
# - stringi [* -> 1.7.6]
# - stringr [* -> 1.4.0]
# - sys [* -> 3.4]
# - tidyr [* -> 1.1.3]
# - tidyverse [* -> 1.3.1]
# - tinytex [* -> 0.36]
# - tzdb [* -> 0.2.0]
# - uuid [* -> 1.0-3]
# - vroom [* -> 1.5.5]
# - xfun [* -> 0.29]
# - xml2 [* -> 1.3.2]
# - yaml [* -> 2.2.1]
#
# Do you want to proceed? [y/N]: y
# * Lockfile written to '~/computational-reproducibility-in-r/renv.lock'.Now, our renv.lock file is updated! Finally, we can go back to a regular terminal in our project directory and add, commit, and push our changes to GitHub:
git add .
git commit -m "adding new dependencies"
git pushWe should be able to navigate to the Actions tab in our GitHub repo and see the image being built in real time!
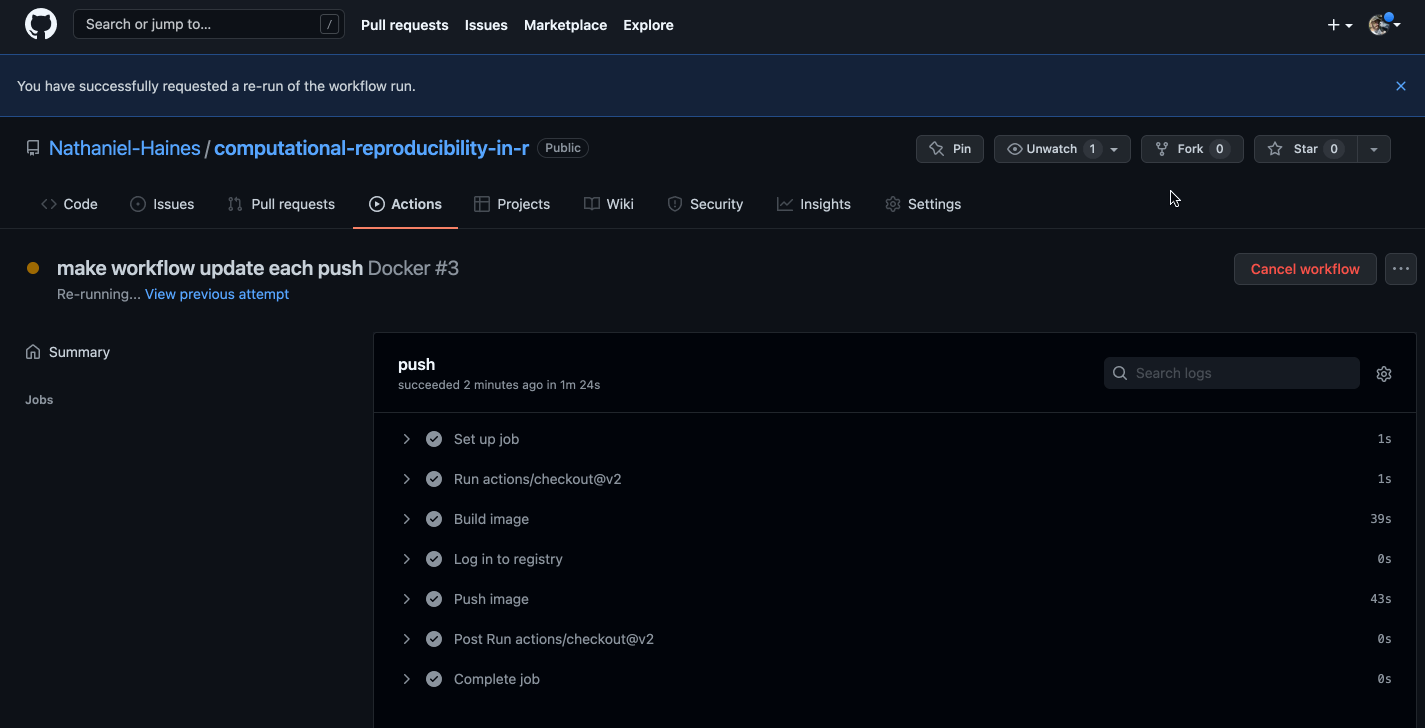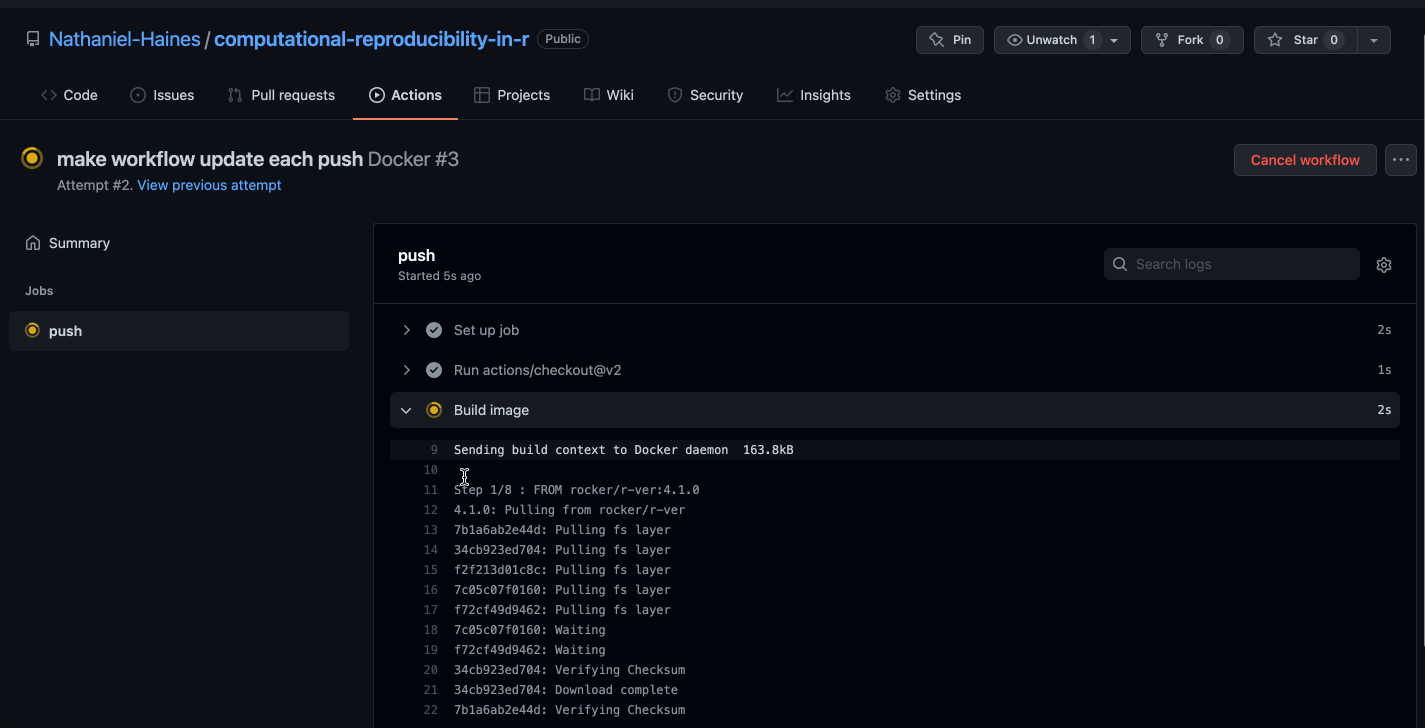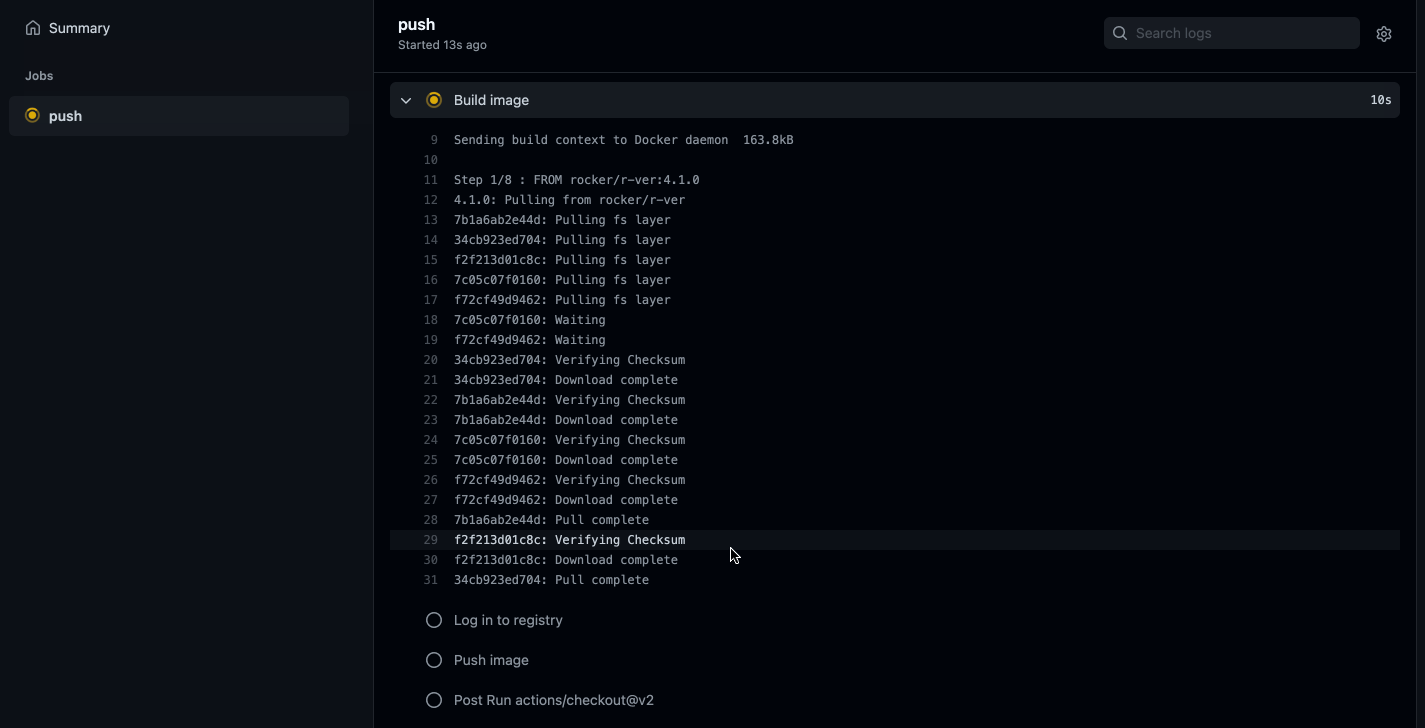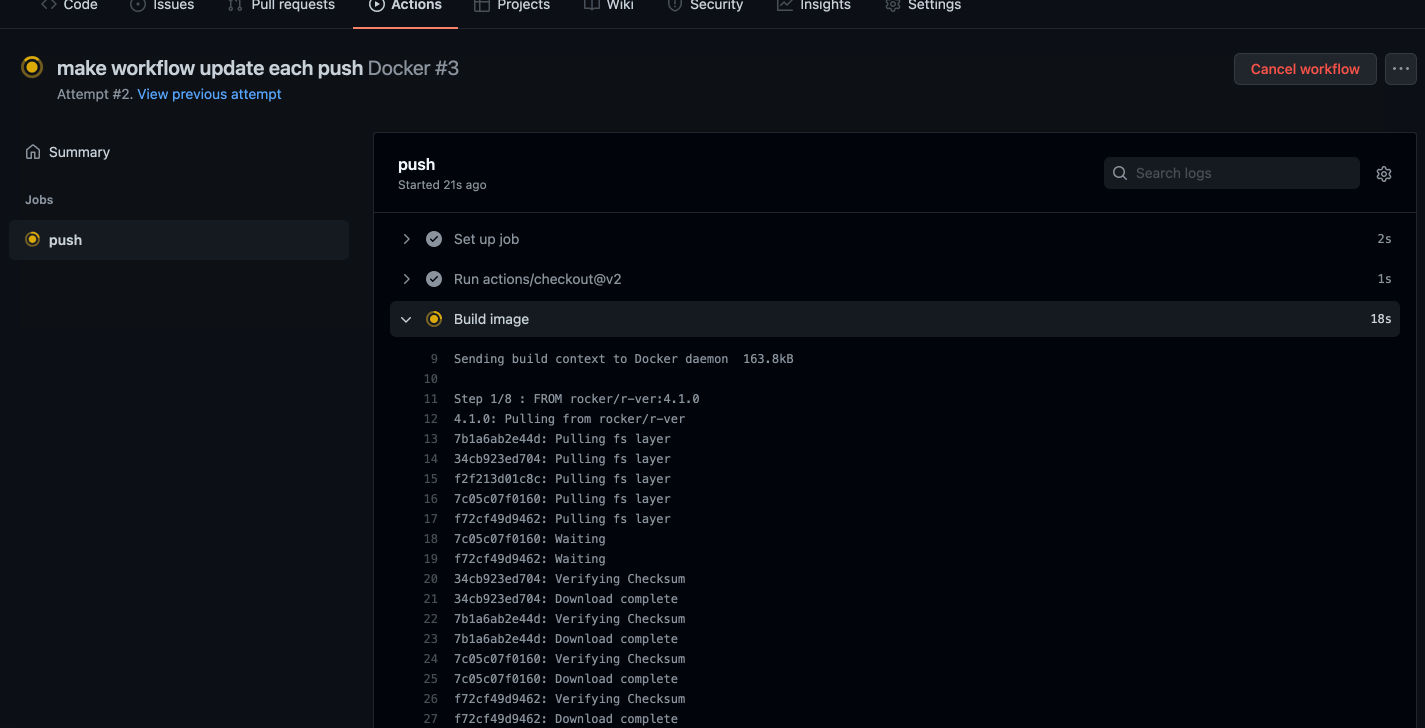
Caption: Well would you look at that, it works!
How to Use Your Docker Images
You will now be able to find you image under the Packages tab on your GitHub profile. For example, you can find the image that my own workflow built here. Here, you will also see a command you can run to download the package so that you (or anyone!) could use the image. In my case, it is: docker pull ghcr.io/nathaniel-haines/computational-reproducibility-in-r:latest. After pulling it to your local computer, you can use it as normal!
Wrapping up
Alright! So that was a bit long, but I hope that it was useful in covering computational reproducibility in R. There are of course many other things you could include in this workflow to make it even better (e.g., only building a new image when the renv.lock file changes, as opposed to each push, or integrating with asdf so that you can use different R versions across local projects akin to how renv lets you use different R package versions), but I will leave that as an exercise for those who are interested :D
Thanks a lot for stopping by, and I hope you find this useful in your work!